Findings on non-coding RNA function
Mechanisms of ncRNA function
Some of our findings regarding the physiological and molecular functions of miRNAs and lncRNAs are summarized in the sections below.
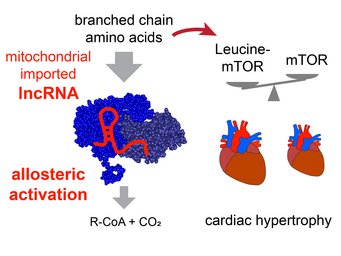
Mitolnc controls BCAA metabolism and heart hypertrophy

Our study reveals that the lncRNA mitolnc (AK079912, GM44386) allosterically activates the enzymatic activity of the branched chain ketoacid dehydrogenase complex. The activity of this lncRNA is needed to prevent accumulation of branched chain amino acids (BCAAs) in cardiac tissue. Increased concentration of the BCAA leucine in cardiomyocytes after inactivation of mitolnc results in mTOR activation and cardiac hypertrophy. For more information look at Weiss et al. (2024) Nucleic Acids Research.
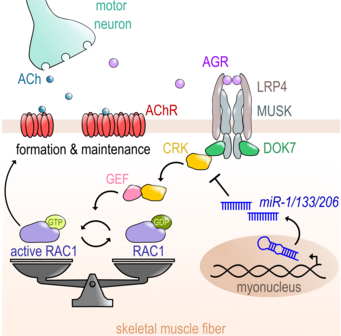
MiR-1/206/133 are essential for neuromuscular junction formation.

Here we demonstrate that in skeletal muscle miR-1/206/133 together are needed to control the expression of an important component of the signalling cascade regulating the formation of neuromuscular junctions (NMJs) of skeletal muscle. Deletion of the miRNAs results in severe disturbance of NMJ formation during development and loss of NMJs in adult live, thereby highlighting the need for tight control of the ubiquitously expressed adapter protein CRK to support NMJ formation in skeletal muscle. For more information look at Klockner et al. (2022) Nature Comm.
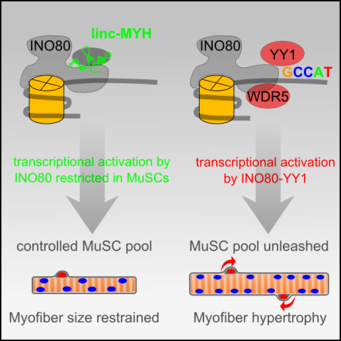
Linc-MYH configures INO80 to regulate muscle stem cell numbers and skeletal muscle hypertrophy

In this work, we reveal that the lncRNA linc-MYH re-configures the ubiquitous chromatin remodeler complex INO80 specifically in proliferating skeletal muscle stem cells (MuSCs) to prevent non-physiological MuSC proliferation and myofiber hypertrophy.
In proliferating MuSCs linc‐MYH interacts in trans directly with the INO80 chromatin remodeler complex and thereby restricts the recruitment of the additional subunits YY1 and WDR5 to the INO80 complex. This reconfiguration controls the function of the INO80 complex in regulation of the transcriptional activity of the YY1 transcription factor without affecting other crucial functions of the ubiquitous INO80 complex in DNA repair. For more information look at Schutt et al. (2020) EMBO J.
Metabolic maturation during muscle stem cell differentiation is achieved by miR-1/133a.

In this work, we have identified a regulatory cascade consisting of the miRNAs miR-1/133a, their direct target MEF2a and the Dlk1-Dio3 megacluster of miRNAs that is essential for the maturation of mitochondria in skeletal muscle cells.
The Dlk1-Dio3 megacluster is the largest cluster of miRNAs in the mammalian genome and is highly expressed in muscle stem cells. Repression of this megacluster by the regulatory cascade is essential to allow metabolic maturation of the skeletal muscle myofibers. Failure of the identified regulatory cascade results in reduced mitochondrial function in skeletal muscle and abrogates endurance running. For more information look at Wüst et al. (2018) Cell Metabolism.
miR-22 controls male muscle lipid metabolism and body weight

The miR-22hg lncRNA (pri-miR-22) containing the miR-22 sequence (blue) is highly expressed striated muscle. In addition, the lncRNA contains a sequence element (green, ERE) resembling the estrogen receptor alpha (ERα) binding sequence known from ERα-DNA binding studies.
Our study reveals, that the release of mature miR-22 from the lncRNA is repressed by binding of ERα to the ERE sequence. Due to differences in ERα activity, the inhibition of miR-22 release is stronger in female that in male skeletal muscle, resulting in higher concentration of active miR-22 in male muscle compared to the female muscle.
On the other hand, miR-22 represses the translation of ERα protein and the abundant miR-22 in male muscle reduces the expression of ERα protein specifically in the male muscle. Reduced ERα activity in male muscle due to this feedback-loop limits the lipid metabolism in male muscle and results in higher body mass of the male animals. In females, the release of active miR-22 is reduced due to ERα activity, thus ERα is not repressed.
Our work identifies a regulatory loop that allows sex- and tissue specific control of ERα function and male-specific regulation of muscle lipid metabolism. For details look at Schweisgut et al. (2017) EMBO J.
The miRNA-clusters miR-1/133a are essential for embryonic heart development

The miRNAs mir-1 and miR-133a are also clustered in the genome. Interestingly, this miRNA cluster appears twice in the genome; in mice on chromosome 2 and chromosome 18. Both miRNA clusters are specifically expressed in heart and muscle and give rise to identical mature miR-1 or miR-133a molecules. Deletion of one or of the other miR-1/133a cluster from the mouse genome does not cause obvious impairment of heart or muscle function in mice; however, deletion of both clusters is embryonic lethal. Using this model, we demonstrated that repression of the transcriptional coactivator myocardin by miR-1 is essential for embryonic heart development. Loss of miR-1 in the mutant mice leads to upregulation of myocardin, which in turn regulates many of the secondary effects seen in miR-1/133a loss of function mice. A main feature of the myocardin gain of function is upregulation of smooth muscle specific genes. The loss of miR-133a in our mutant animals reveals that miR-133a represses the smooth muscle specific potassium channel Kcnmb1. Transgenic overexpression of myocardin phenocopies many aspects of the miR-1/133a mutant phenotype by activation of the smooth muscle gene program in the embryonic heart. Indeed myocardin also activates transcription of Kcnmb1.
Our data reveal that miR-1/133a is essential to repress a myocardin-induced smooth muscle gene program in the embryonic heart to allow further cardiomyocyte development. In addition myocardin activates the expression of the miR-1/133a clusters, thus our work proves a regulatory feedback-loop between miR-1/133a, myocardin and Kcnmb1 that is essential for early heart development. For details look at Wystub et al. (2013) PLOS Genetics.
The miR-143/145 cluster is essential for the contractile phenotype of VSMCs

The miRNAs miR-143 and miR-145 are highly expressed in smooth muscle cells. In vascular smooth muscle cells (VSMCs) these miRNAs repress the expression of target proteins like angiotensin converting enzyme (ACE). Repression of ACE is essential for VSMCs to prevent the local release of angiotensin II from angiotensin I. Loss of the ACE-repression in miR-143/145 mutant mice leads to ectopic expression of ACE in VSMCs and thus excessive stimulation of VSMCs by the agonist angiotensin II. This leads to loss of agonist-induced contractility of VSMCs, reduced blood pressure, a synthetic phenotype of VSMCs and arteriosclerosis. Systemic inhibition of ACE partially restores the VSMC phenotype, indicating that ACE is an important target of miR-143/145, that is essential for maintenance of contractility and the contractile phenotype of VSMCs. In addition to ACE, there are also other mechanisms of miR-143/145 function that need to be explored. For details look at Boettger et al. (2009) JCI.